Use Experience
Scientists have learned to rely on the grid as a reliable and inexpensive way to process massive amounts of data. Starting from this common experience, which also involves existing computer centers, Auger and WeNMR over WNoDeS, are intended to provide grid and cloud services based on the proven reliability of the batch system and overcome its limitations in terms of simplicity of access and configuration for the users, pre-packing the software and configuration over virtual machine images.
INFN Tier-1
The INFN Tier-1 computing centre (CNAF, Bologna) manages a WNoDeS production instance on top of the IBM/Platform LSF batch system and IBM GPFS storage disk since November 2009. It works in a fully integrated fashion with the overall number of 13 thousand cores of the farm, all belonging to a single LSF cluster. At the time of this writing, almost one thousand VMs can concurrently be instantiated on-demand out of this common farm to support cloud computing, or custom executing environments.
At INFN Tier-1 a certain number of Virtual Organizations (VOs) have been configured to use WNoDeS during the computation of their jobs. VOs supported at INFN Tier-1 computing centre using WNoDeS are alice, ams, biomed, cms, lhcb, pamela and of course auger_db, enmr.eu. In the FedCloud context the WNoDeS deployment also supports: dteam, fedcloud, gridit, ops, testers.eu-emi. Furthermore, the site adopts mixed mode to use physical resources both as traditional batch nodes and, at the same time, as HVs for VM instantiation. This lets sites to introduce features like the support of VM and cloud computing on traditional resources without disrupting existing services and allowing to efficiently deciding which workloads are to be virtualized and which should be run instead on top of non-virtual hardware.
With the mixed mode turned on, any node of a farm may act as a traditional, "real" node (that only runs traditional batch jobs), as a pure hypervisor (that only runs VMs), or as a node able of running, at the same time, both jobs on the physical hardware (without thus incurring penalties typical of virtual infrastructures), and jobs or cloud services on VMs (thus exploiting capabilities offered by virtual environments). Therefore, mixed mode greatly enhances the flexibility of the configuration of a farm and lets system administrators progressively integrated WNoDeS into existing data centres.
Astro-Particle Physics Community
The Pierre Auger Cosmic Ray Observatory [1] is a cosmic ray observatory located in Argentina that studies ultra-high energy cosmic rays, the most energetic and rarest of particles in the universe. Scientists involved in the Pierre Auger Observatory deal with so far unknown sources of cosmic rays that impact our planet. When these particles strike the earth's atmosphere, they produce extensive air showers made of billions of secondary particles. While much progress has been made in nearly a century of research in understanding cosmic rays with low to moderate energies, those with extremely high energies remain mysterious. These rays have energy many times greater than what we can achieve in our largest particle accelerators. The scientists measure showers of particles caused by cosmic rays using detectors deployed across three thousand square kilometers of the Pampas of Argentina: they require massive computational power for simulation to compare readings with theoretical models.
The Auger's computational model requires concurrent read only access to a condition data base (MySQL) from hundreds of computing nodes in the farm. The data base resides in a networked filesystem (IBM/GPFS) and may be concurrently accessed by one or more MySQL engines. This of course is only possible in the case of read only access. Being a single database engine ineffective at serving hundreds of clients, an opposite solution was initially preferred by Auger: a database engine would have been available at every node, each one independently accessing the filesystem with its data base. This approach however lead again to poor performances because of the high rate of non-serialized and concurrent disk accesses that causes overload on the GPFS side.
A sustainable solution has been successfully implemented at INFN Tier-1, by running the Auger's jobs on Virtual WNs managed by WNoDeS. Each one of these selects and retrieves its needed data from a MySQL engine installed on the Hypervisor where the virtual WN have been instantiated. In so doing, the number of needed database engines is reduced by a factor equal to the number of concurrent runnable jobs in a physical host, which in turn equals the number of cores of the hypervisor. To further optimize this solution, a job packing configuration [2] has been recently investigated and tested, and is being applied to the LSF batch system of the INFN Tier-1. This is done to have the Auger's "virtual" jobs being concentrated on the smallest possible set of physical nodes, thus optimally reducing the number of running MySQL engines.
Life Science with WeNMR Virtual Research Community
WeNMR [3] is both a EU FP7 funded project and a Virtual Research Community (VRC) supporting the Nuclear Magnetic Resonance (NMR) and Small Angle X-ray Scattering (SAXS) structural biology user community. It currently operates the largest VO in life science domain of the European Grid Infrastructure (EGI), with more than 500 registered users worldwide.
While mainly offering application portals providing 'protocolized' access to EGI grid, one of their use-case better fits with the cloud model. The Common Interface for NMR Structure Generation (CING [4]) is a software suite designed for the residue-based validation of bio-molecular three-dimensional structures determined by NMR. The CING framework integrates about 20 different external programs and additionally uses internal routines to validate NMR-derived structure ensembles against empirical data and measured chemical shifts, distance and dihedral restraints. Due to the high number of external dependencies CING cannot easily be installed on traditional grid computing nodes. In contrast, the collection of programs could easily be packaged into a virtual machine that we called "VirtualCING". Therefore, VirtualCING is very suited to be used in a cloud environment. For example, it has been and still is successfully applied to establish the NRG-CING database, an annotated and re-mediated repository of experimental, structural, and validation data [5]. The import of one NMR structure into the database and the creation of a CING validation report on average require 20 minutes on a single core, taking about 3,300 core hours to process the current set of entries.
The experimental and computational procedures involved in the determination of biomolecular structures by NMR are continuously being developed and improved, and have become more advanced over the past 25 years. Therefore, by applying today's improved technology to the original data, better structures can be calculated [6,7]. Recently, we extended VirtualCING with routines for the recalculation of NMR ensembles using the experimental data in the NRG-CING repository, allowing us to extend our previous recalculation projects and re-determine NMR structures in the Protein Databank (PDB) [8] in the cloud on a large scale. The NMR_REDO project aims to improve quality of NMR-derived protein structure ensembles in terms of fit with the original experimental data and geometric quality. To date, 3,400 structures have been recalculated and we expect this number to increase to 5,000 of the 10,000 structures deposited in the PDB when our protocol can handle all usable deposited experimental data. A full NMR_REDO iteration currently consumes about 25,000 core hours. Preliminary results indicate that the recomputed ensembles generally show a better fit to the original data and to independent validation criteria than the original ensembles.
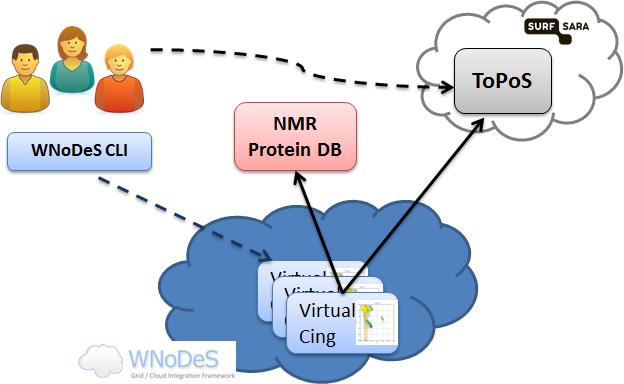
Figure 1: Architecture of the WNoDeS-based cloud infrastructure for the WeNMR/VirtualCING use case.
To verify how the WNoDeS cloud framework can enable scientists to perform NMR computations, a customised VirtualCING image has been deployed in the WNoDeS marketplace. The WeNMR user, after having created his own VOMS proxy certificate, instantiates a number of VirtualCING machines through the WNoDeS CLI. After booting, each VirtualCING automatically starts a process getting the job payload from the ToPoS token pool server, a pilot job framework offered through a HTTP server hosted by SURFsara organization, in The Netherlands. The tokens, previously uploaded on the ToPoS server by the user, contain: the information about the proteins to be processed, the URL of the input data to be fetched from the NRG-CING Protein DB, the location of the NMR_REDO Protein DB where to upload the final output data (both DBs are represented by the pink box in Figure 1), and a set of parameters as arguments of the executable. At the end of the computation the token is deleted from the ToPoS server and the VirtualCING process asks for another unlocked token, until all tokens have been processed and nothing is left on the ToPoS server. The final results, for each recalculated protein, can be then visualized through the web interface of the NMR_REDO Protein DB.
References
[1] Pierre Auger Observatory project http://www.auger.org.
[2] Dal Pra S., "Job Packing: optimized configuration for job scheduling", presentation at HEPIX 2013, 15-19 April 2013, Bologna, Italy.
[3] Wassenaar et al. (2012), "WeNMR: Structural Biology on the Grid", J. Grid. Comp., 10:743-767
[4] Doreleijers, J. F. et al., "CING: an integrated residue-based structure validation program suite", Journal of biomolecular NMR (2012). doi:10.1007/s10858-012-9669-7.
[5] Doreleijers, J. F. et al., "NRG-CING: integrated validation reports of remediated experimental biomolecular NMR data and coordinates in (wwPDB)", Nucleic acids research 40, D519-24 (2012).
[6] Nabuurs, S. B. et al., "DRESS: a database of refined solution NMR structures", Proteins 55, 483-6 (2004).
[7] Nederveen, A. J. et al., "RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank", Proteins 59, 662-72 (2005).
[8] Berman, H., Henrick, K., Nakamura, H. & Markley, J. L., "The worldwide Protein Data Bank wwPDB: ensuring a single, uniform archive of PDB data", Nucleic acids research 35, D301-3 (2007).